Complex traits genetics
Complex trait genetics explores how numerous genetic variants, often of small individual effect, interact with each other and with the environment to influence multifaceted phenotypes such as growth, reproduction, and disease resistance. In our work, we apply quantitative genetic appraoches (such as genome-wide association study [GWAS], molecular QTL mapping), and integrative multi-omics to dissect the genetic architecture of complex traits and diseases. By linking variation in DNA sequence to changes in molecualr phenotypes, regulatory networks, and phenotypic outcomes, we aim to identify causal loci, reveal underlying biological mechanisms, and provide a predictive framework for breeding and biomedicine.
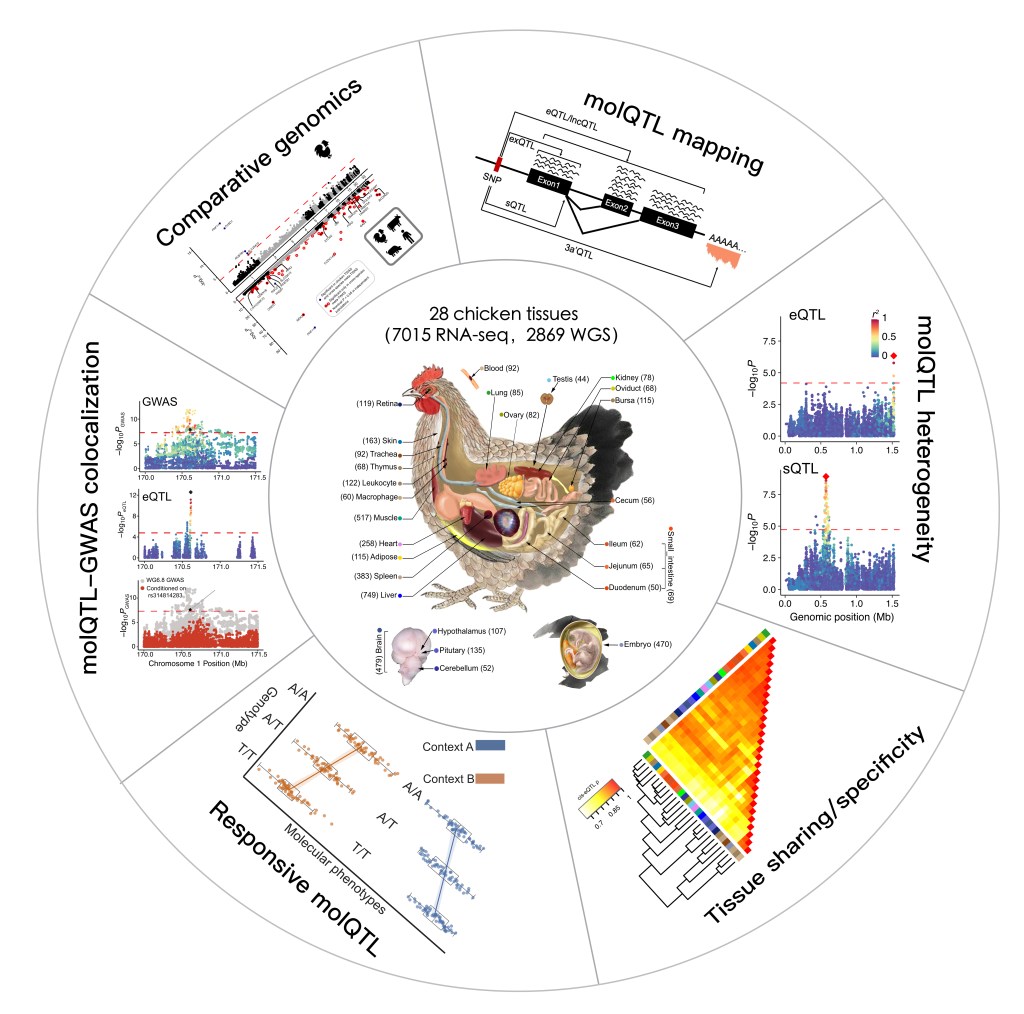

Comparative genomics
Comparative genomics investigates how regulatory landscapes evolve and diversify across species. In our research, we generate and integrate multi-tissue epigenomic datasets—such as chromatin accessibility, histone modifications, and transcriptomic profiles—from animals through initiatives like ChickenGTEx and FAANG and from human through initiatives like ENCODE and RoadMap Epigneomcis. By comparing these regulatory maps across species, we identify conserved elements and lineage-specific innovations, revealing how evolution, selection, and demographic history shape genome function. This approach provides critical insights into the mechanisms that link genetic variation to phenotypic diversity and evolutionary adaptation.
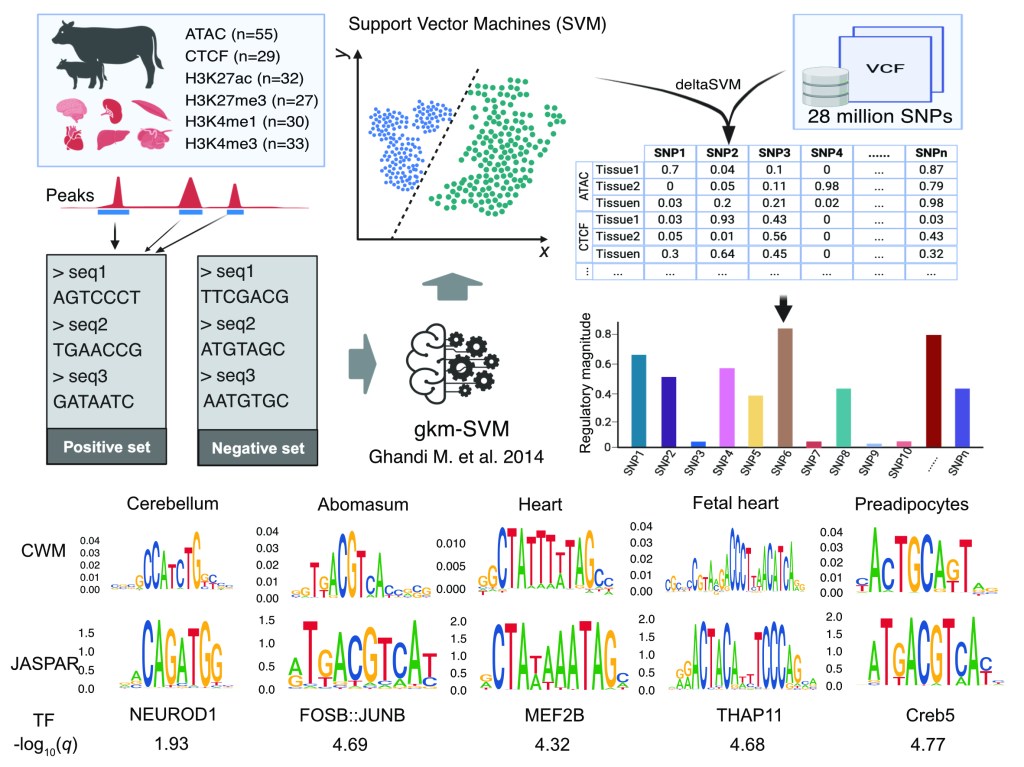
Sequence-to-Function
Sequence-to-function modeling uses computational and statistical frameworks to predict how DNA sequence variation influences molecular and organismal phenotypes. In our research, we develop and apply deep-learning and integrative genomics approaches—building on large datasets from ChickenGTEx, FAANG, ENCODE and other resources—to infer regulatory effects directly from primary DNA sequence. These models enable “zero-shot” prediction of gene regulation and variant effects across previously uncharacterized populations and species, bridging the gap between sequence variation, regulatory mechanisms, and complex traits. By doing so, we create powerful tools for uncovering causal variants, prioritizing functional regions, and guiding precision breeding strategies.